INDELible
[MODEL] block
- These blocks are used to specify the evolutionary models used during simulation.
- There are a variety of commands available for specifying different substitution models and indel models as well as rates of insertion and deletion to substitution etc.
- Each model block in the control file must be given a name (e.g. "mymodelname" below). This is used to refer to the model elsewhere in the control file.
- It does not matter whether the "white space" (between commands and values) is spaces, tabs or new lines.
- If a command is not specified the default value is used. Default values are listed in the description of each command.
- Please click on any of the commands to learn more about them.
Example Usage:
[MODEL] mymodelname [submodel] HKY 2.5 // HKY with kappa of 2.5 [indelmodel] LAV 1.7 541 // specifies the indel length distribution [indelrate] 0.1 // rates of insertion and deletion are both 0.1 [geneticcode] 2 // only used in CODON simulations [rates] 0.25 0.50 10 // pinv alpha ngamcat [statefreq] 0.4 0.3 0.2 0.1 // frequencies for T C A G
[submodel] for NUCLEOTIDE simulations
- Follow the links for information about this command for AMINOACID or CODON simulations.
- Most substitution models implemented in INDELible are variations of the following general matrix:
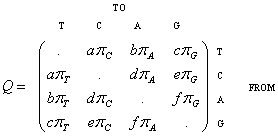
+-----+----------------------------------+----------------------------------+ | N | Usage | Notes | +-----+----------------------------------+----------------------------------+ | 0 | [submodel] JC | a=b=c=d=e=f=1 | | 1 | [submodel] F81 | a=b=c=d=e=f=1 | +-----+----------------------------------+----------------------------------+ | 2 | [submodel] K80 a | a=f=kappa, b=c=d=e=1 | | 3 | [submodel] HKY a | a=f=kappa, b=c=d=e=1 | +-----+----------------------------------+----------------------------------+ | 4 | [submodel] TrNef a f | a=kappa1, f=kappa2, b=c=d=e=1 | | 5 | [submodel] TrN a f | a=kappa1, f=kappa2, b=c=d=e=1 | +-----+----------------------------------+----------------------------------+ | 6 | [submodel] K81 b c | b=e, c=d, a=f=1 | | 7 | [submodel] K81uf b c | b=e, c=d, a=f=1 | +-----+----------------------------------+----------------------------------+ | 8 | [submodel] TIMef a b c | b=e, c=d, f=1 | | 9 | [submodel] TIM a b c | b=e, c=d, f=1 | +-----+----------------------------------+----------------------------------+ | 10 | [submodel] TVMef b c d e | a=f=1 | | 11 | [submodel] TVM b c d e | a=f=1 | +-----+----------------------------------+----------------------------------+ | 12 | [submodel] SYM a b c d e | f=1 | | 13 | [submodel] GTR a b c d e | f=1 | +-----+----------------------------------+----------------------------------+ | 14 | [submodel] F84ef k | b=c=d=e=1, a=(1+k/Y), f=(1+k/R) | | 15 | [submodel] F84 k | b=c=d=e=1, a=(1+k/Y), f=(1+k/R) | +-----+----------------------------------+--------------------------+-------+ | 16 | [submodel] UNREST TC TA TG CT CA CG AT AC AG GT GC | GA=1 | +-----+-------------------------------------------------------------+-------+ N.B. N can be substituted for the model name. e.g. [submodel] 0 instead of [submodel] JC
[submodel] for AMINOACID simulations
- Follow the links for information about this command for NUCLEOTIDE or CODON simulations.
- This command is quite simple in this case. Usage is just: [submodel] value
- value is just an integer N or a code used to pick the amino-acid substitution model as defined below (references from paper given where appropriate):
+-----+---------------+-------------------------------+ | N | code | Reference | +-----+---------------+-------------------------------+ | 0 | Poisson | n/a | | 1 | JTT | Jones et al,. 1992 | | 2 | JTT-dcmut | Kosiol and Goldman, 2005 | | 3 | Dayhoff | Dayhoff et al., 1978 | | 4 | Dayhoff-dcmut | Kosiol and Goldman, 2005 | | 5 | WAG | Whelan and Goldman, 2001 | | 6 | mtMAM | Yang et al., 1998 | | 7 | mtART | Abascal et al., 2007 | | 8 | mtREV | Adachi and Hasegawa, 1996 | | 9 | rtREV | Dimmic et al., 2002 | | 10 | cpREV | Adachi, 2000 | | 11 | Vt | Muller and Vingron, 2000 | | 12 | Blosum | Henikoff and Henikoff, 1992 | | 13 | LG | Le and Gascuel, 2008 | | 14 | HIVb | Nickle et al., 2007 | | 15 | HIVw | Nickle et al., 2007 | | 16 | USER | n/a | +-----+---------------+-------------------------------+
[submodel] USER userAAmodel.txt
[submodel] for CODON simulations
- Follow the links for information about this command for NUCLEOTIDE or AMINOACID simulations.
- This command is [submodel] ECMunrest for the empirical unrestricted model.
- This command is [submodel] ECMrest for the empirical restricted model.
- Otherwise a codon model with K discrete omega categories is defined using the commands below in blue:
// M3 (discrete) // p(K-1)=1-p(K-2)-...-p1-p0
[submodel] kappa
p0 p1 ... p(K-2) // proportions
ω0 ω1 ... ω(K-2) ω(K-1) // omegas
// M0 (one-ratio)
[submodel] kappa &omega0 // p0=1
// M1 (neutral)
[submodel] kappa p0 ω0 1 // ω1=1; p1=1-p0
// M2 (selection)
[submodel] kappa p0 p1 ω0 1 ω2 // ω1=1; p2=1-p1-p0
// M4 (freqs) with K=5
[submodel] kappa
p0 p1 p2 p3 // p4=1-p3-p2-p1-p0
0 0.333333 0.666666 1 3 // ω0, ω1, ω2, ω3, ω4
[indelmodel]
- This sets the insertion and deletion length distributions to be the same with four possible choices:
(1) [indelmodel] NB q r // Negative Binomial Distribution (2a) [indelmodel] POW a // Zipfian Distribution (2b) [indelmodel] POW a M // Zipfian Distribution (3) [indelmodel] LAV a M // Lavalette Distribution (4) [indelmodel] USER mylengthmodel.txt // User-Defined Distribution
[indelrate]
- This sets the insertion and deletion rates to both be equal to whatever value is given.
- Both rates are relative to an average substitution rate of 1.
- N.B. This means that insertionrate = indelrate and deletionrate = indelrate.
- This is not the same as insertionrate + deletionrate = indelrate.
- To specify different rates for insertions and deletions then do not use [indelrate].
- In this case use the commands [insertrate] and [deleterate] instead. The values for each command are the rates relative to an average substitution rate of 1.
- Return to example usage.
[geneticcode]
- This command can only be used in CODON simulations.
- The value should be an integer 1 to 6, 9 to 16, or 21 to 24, corresponding to the genetic codes listed on Genbank.
- The value 1 (corresponding to the universal genetic code) is the default setting if the command is not specified.
- These genetic codes determine which codons are stop codons and therefore not simulated by INDELible.
- They are also used to translate codons to amino-acids for output if that option is chosen.
- The codes listed at Genbank (in Oct. 2008) are given below (* represents a stop codon).
- Please note some codes are identical and differ only in terms of Starts. Please see Genbank for more info.
1 - The Standard Code
FFLLSSSSYY**CC*WLLLLPPPPHHQQRRRRIIIMTTTTNNKKSSRRVVVVAAAADDEEGGGG
2 - The Vertebrate Mitochondrial Code
FFLLSSSSYY**CCWWLLLLPPPPHHQQRRRRIIMMTTTTNNKKSS**VVVVAAAADDEEGGGG
3 - The Yeast Mitochondrial Code
FFLLSSSSYY**CCWWTTTTPPPPHHQQRRRRIIMMTTTTNNKKSSRRVVVVAAAADDEEGGGG
4 - The Mold, Protozoan, and Coelenterate Mitochondrial
Code and the Mycoplasma/Spiroplasma Code
FFLLSSSSYY**CCWWLLLLPPPPHHQQRRRRIIIMTTTTNNKKSSRRVVVVAAAADDEEGGGG
5 - The Invertebrate Mitochondrial Code
FFLLSSSSYY**CCWWLLLLPPPPHHQQRRRRIIMMTTTTNNKKSSSSVVVVAAAADDEEGGGG
6 - The Ciliate, Dasycladacean and Hexamita Nuclear Code
FFLLSSSSYYQQCC*WLLLLPPPPHHQQRRRRIIIMTTTTNNKKSSRRVVVVAAAADDEEGGGG
9 - The Echinoderm and Flatworm Mitochondrial Code
FFLLSSSSYY**CCWWLLLLPPPPHHQQRRRRIIIMTTTTNNNKSSSSVVVVAAAADDEEGGGG
10 - The Euplotid Nuclear Code
FFLLSSSSYY**CCCWLLLLPPPPHHQQRRRRIIIMTTTTNNKKSSRRVVVVAAAADDEEGGGG
11 - The Bacterial and Plant Plastid Code
FFLLSSSSYY**CC*WLLLLPPPPHHQQRRRRIIIMTTTTNNKKSSRRVVVVAAAADDEEGGGG
12 - The Alternative Yeast Nuclear Code
FFLLSSSSYY**CC*WLLLSPPPPHHQQRRRRIIIMTTTTNNKKSSRRVVVVAAAADDEEGGGG
13 - The Ascidian Mitochondrial Code
FFLLSSSSYY**CCWWLLLLPPPPHHQQRRRRIIMMTTTTNNKKSSGGVVVVAAAADDEEGGGG
14 - The Alternative Flatworm Mitochondrial Code
FFLLSSSSYYY*CCWWLLLLPPPPHHQQRRRRIIIMTTTTNNNKSSSSVVVVAAAADDEEGGGG
15 - The Blepharisma Nuclear Code
FFLLSSSSYY*QCC*WLLLLPPPPHHQQRRRRIIIMTTTTNNKKSSRRVVVVAAAADDEEGGGG
16 - The Chlorophycean Mitochondrial Code
FFLLSSSSYY*LCC*WLLLLPPPPHHQQRRRRIIIMTTTTNNKKSSRRVVVVAAAADDEEGGGG
21 - The Trematode Mitochondrial Code
FFLLSSSSYY**CCWWLLLLPPPPHHQQRRRRIIMMTTTTNNNKSSSSVVVVAAAADDEEGGGG
22 - The Scenedesmus obliquus mitochondrial Code
FFLLSS*SYY*LCC*WLLLLPPPPHHQQRRRRIIIMTTTTNNKKSSRRVVVVAAAADDEEGGGG
23 - The Thraustochytrium Mitochondrial Code
FF*LSSSSYY**CC*WLLLLPPPPHHQQRRRRIIIMTTTTNNKKSSRRVVVVAAAADDEEGGGG
Base1: TTTTTTTTTTTTTTTTCCCCCCCCCCCCCCCCAAAAAAAAAAAAAAAAGGGGGGGGGGGGGGGG
Base2: TTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGG
Base3: TCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAG
[rates]
- This command has no effect in CODON simulations.
- The 3 entries in this command are (from left to right): pinv, alpha and ngamcat.
- pinv is the proportion of invariable sites (should be a number between 0 and 1).
- alpha is the shape parameter for the gamma distribution (should be a positive number).
- If alpha=0 then there will be no gamma rate variation.
- ngamcat is the number of categories to use in the discrete gamma approximation.
- If ngamcat=0 then continuous gamma distribution will be used for rate variation.
- Return to example usage.
[statefreq]
- This command is used to specify the stationary frequencies used in the model by listing them separated by white space.
- If the command is not specified in a [MODEL] block then all stationary frequencies will be set to be equal.
- If the list of numbers does not add up to 1 then they will be rescaled so that they do.
- For NUCLEOTIDE simulations this must be a list of 4 numbers representing the frequencies for the different nucleotides (in the order T C A G).
- For AMINOACID simulations this must be a list of 20 numbers representing the frequencies for the different amino-acids (in the order A R N D C Q E G H I L K M F P S T W Y V). This will change the stationary frequencies from those defined in by the substitution model specified in [submodel] (i.e. INDELible will simulate under a +F variant).
- For CODON simulations this must be a list of 64 numbers representing the frequencies for the different codons (in the order TTT TTC TTA TTG TCT TCC TCA .... GGA GGG).
- Care should be taken that the stationary frequencies corresponding to any stop codons in your chosen genetic code are equal to zero. If your input contradicts this then INDELible will inform you.
- Return to example usage.
